Empirical Risk Loss
In the Gradient Descent blog post, we computed the empirical risk equation: \[L(w) = \frac{1}{n}\sum_{i=1}^{n} \ell({\langle w, x_{i} \rangle, y_{i})}\]
where the logistic loss was: \[ \ell({\hat{y}, y}) = -y * log(\sigma(\hat{y})) - (1-y)* log(1-\sigma(\hat{y})) \]
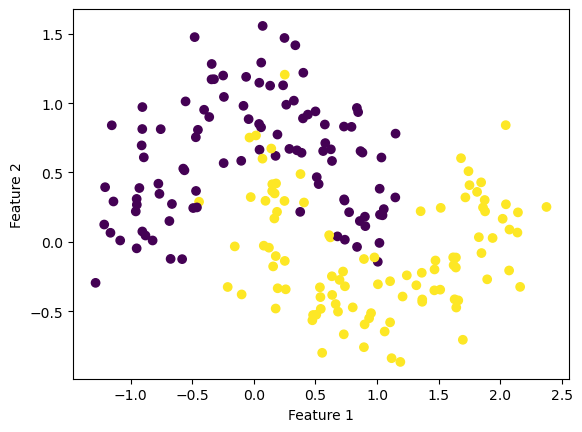
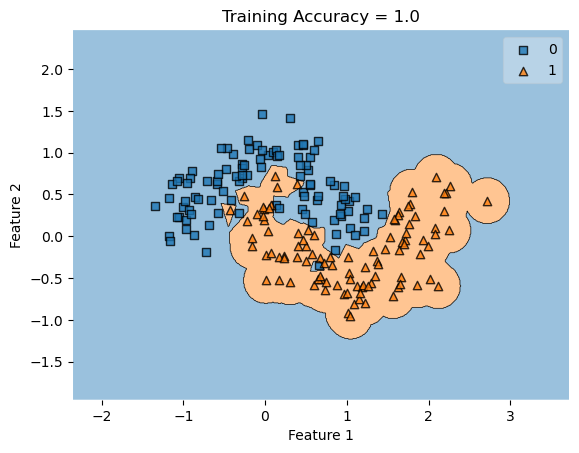
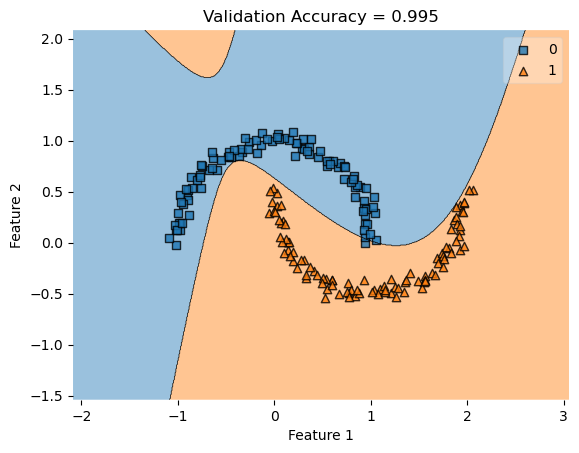
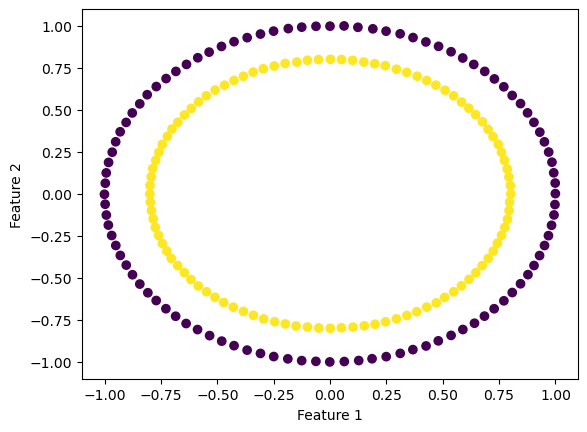
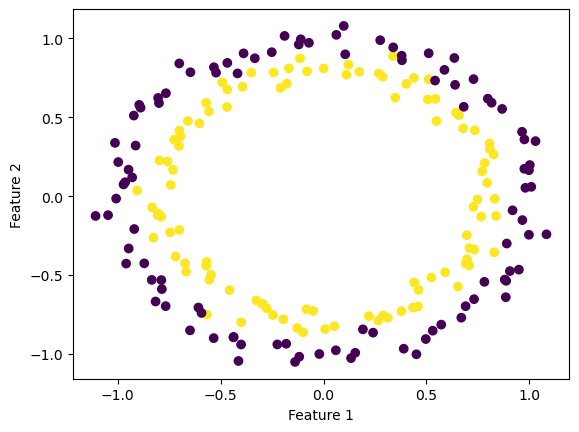
For the kernel logistic regression, by using the kernel trick, we are able to modify our feature matrix \(X\) to be of infinite-dimensional. This means that by transforming our feature matrix X to a kernel matrix, we are able to extend the binary classification of logistic regression for nonlinear features.
Now, the empirical risk for the kernel logistic regression looks like:
\[L(w) = \frac{1}{n}\sum_{i=1}^{n} \ell({\langle v, \kappa({x_{i}}) \rangle, y_{i})}\]
where \(\kappa({x_{i}})\) represents the modified feature vector with row dimension \(\in{R^n}\).
To translate the new empirical risk for the kernel logistic regression into code, I modified my empirical_risk method to take in additional parameters \(km\) and \(w\); \(km\) represents the kernel matrix, and \(w\) represents the weight vector to be optimized. Additionally, I modified my logistic loss method such that it now takes in a pre-computed y_hat as a parameter (originally, the method had taken in a y_hat using the predict method).
We use this loss method to compute the empirical risk by using the inner product between \(km\) and our weight vector \(w\) as our y_hat. Finally, we can take the mean of the loss of the inner product and the true label y to generate our overall empirical risk loss.
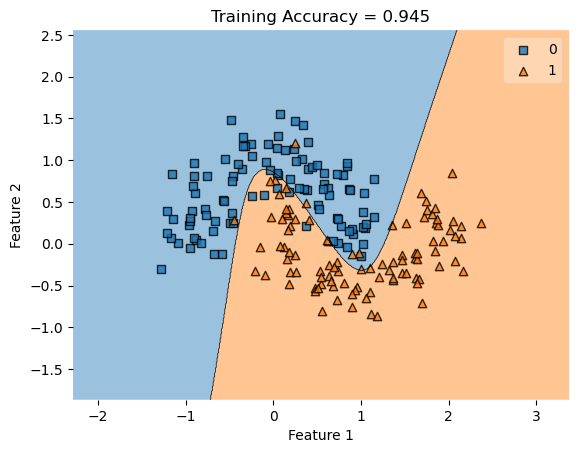
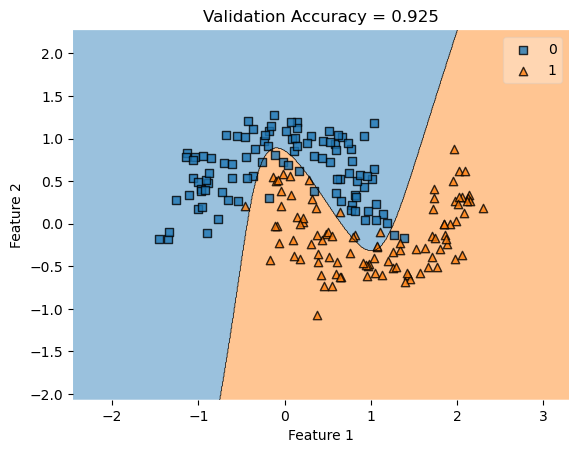
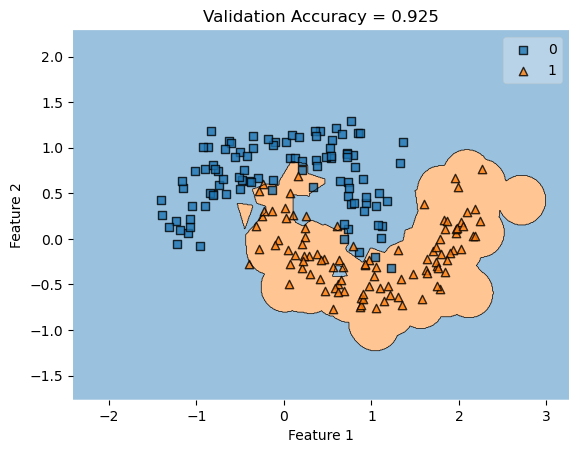
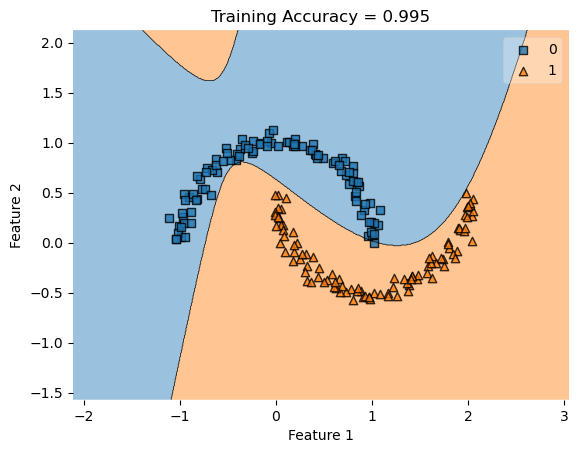
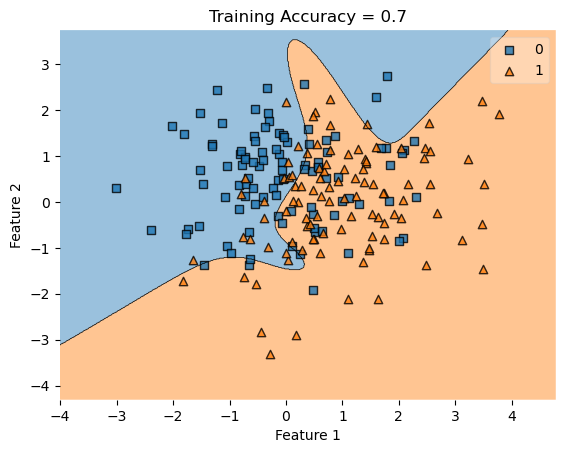
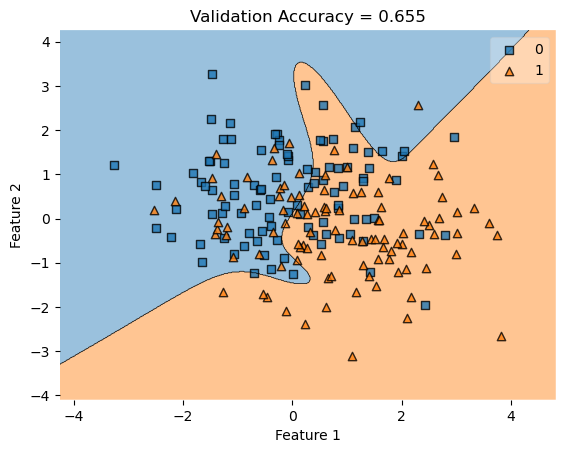
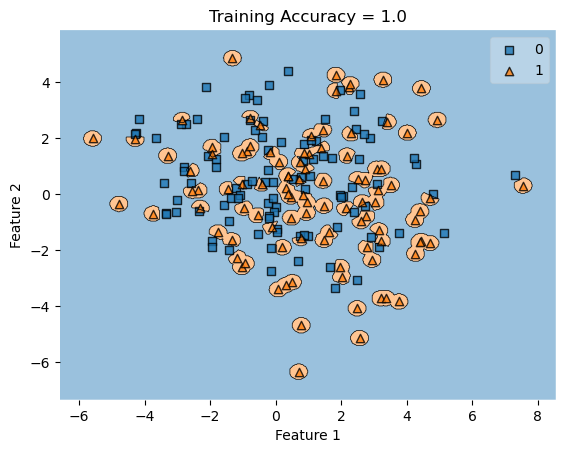
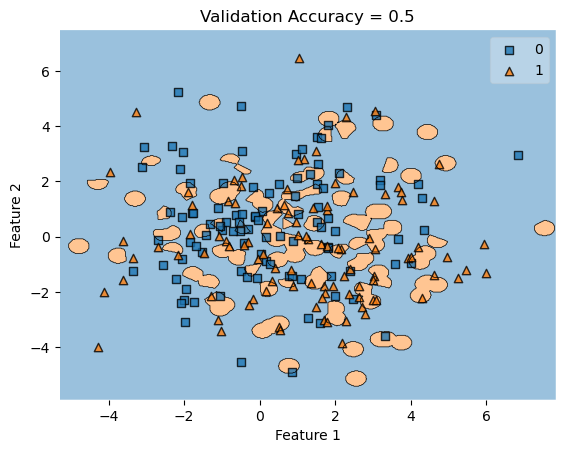
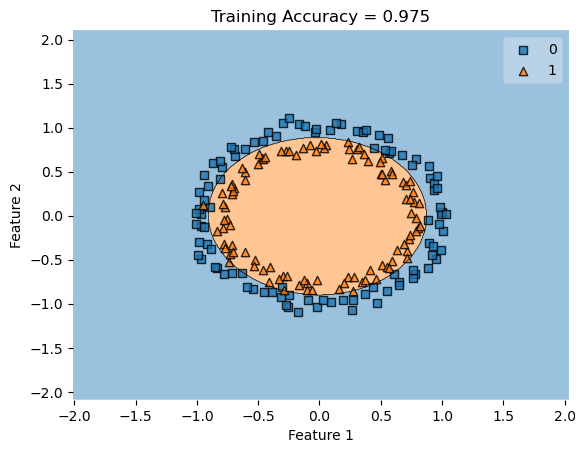
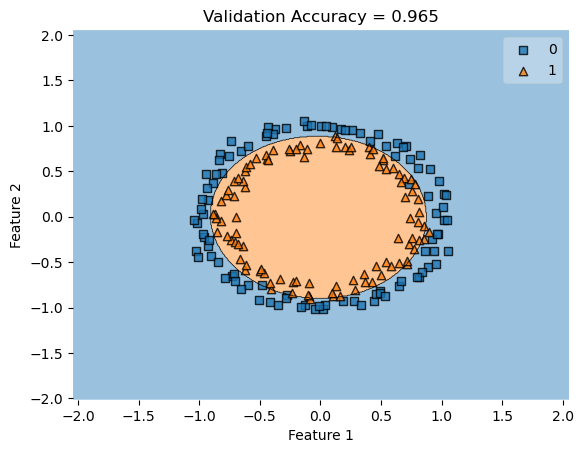
In my fit method, one challenge I faced was the structure of the optimized \(w\) weight vector. In this implementation, I had used the function scipy.optimize.minimize() to optimize my \(w\); the optimized \(w\), however, continuously generated a vector of positive values. This would cause an error in my experiments as the plot_decision_regions function will not be able to plot the trained model’s score.
To fix this problem, after initializing some initial vector \(w_0\) of dimension \(X.shape[0]\), I subtracted \(w_0\) by \(0.5\) to generate both negative and positive values in the optimized \(w\) weight vector.